About Me

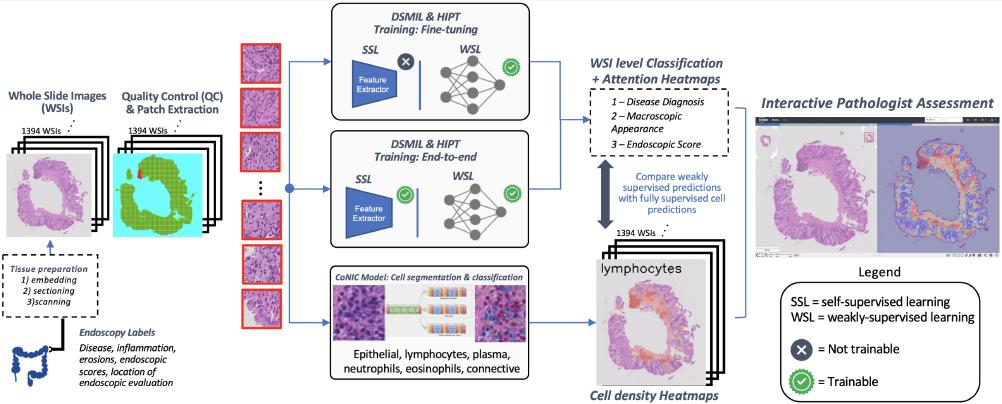
Hi, I’m Ricardo. I am currently an AI Research Scientist at AstraZeneca in Cambridge, UK. At AstraZeneca I have focused on delivering disease insights using state of the art Machine Learning approaches to genomic and medical imaging data, with a focus on model explainability. My most recent work includes developing interpretable Computer Vision models for predicting disease relevant features from images of tissues using self-supervised learning and vision transformers (currently accepted for publication at MIDL).
Prior to joining AstraZeneca, I graduated from Imperial College London with a 1st Class (Dean’s List) MEng degree in Molecular Bioengineering. During my time at Imperial I was a part of the Biological Control Systems Lab, led by Dr. Reiko Tanaka, where I researched the use of state of the art generative models (StyleGAN, Pix2Pix, VAEs) as a data augmentation technique for improving the predictive performance and adversarial robustness of deep Computer Vision models. I was also a part of the Advanced Data Science Team, where I worked on an industrial research project with Refinitiv on autonomous web crawling using Reinforcement Learning.
In my free time I hugely enjoy Sci Fi (books, movies and video games), books about Biology and Theoretical Physics (right now reading Nick Lane and Jim Al-Khalili), weight lifting and swimming.